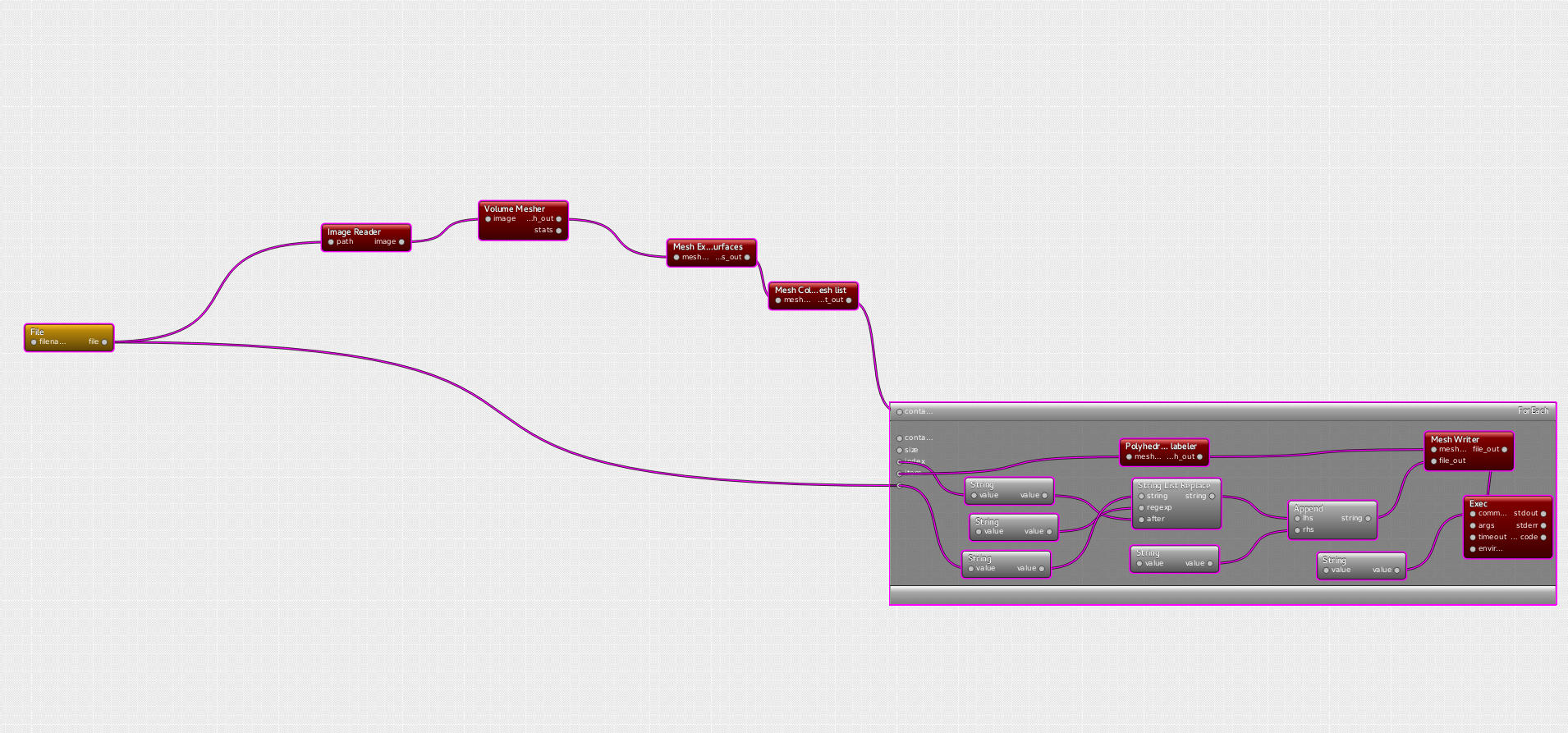
Visual programming framework of dtk enables to create very complex pipelines in a very short time.
The case at stake here is provided by provided by Shuman Jia, PhD at Asclepios and it involves image processing algorithms and discrete geometry.
Heart Surface Extraction pipeline
The purpose of this pipeline is to generate a triangulated surface mesh representing the external surface of a heart, from a mask given as a ".mha" image. Then, from this surface mesh, one wants automatically carrying out a segmentation to isolate the main features of the heart: ventricule, superior vena cava/ ascending aorta, inferior vena cava/descending aorta, and the aortic arch.
The constraint for the mesher is a relatively small number of triangles (<30k) while keeping both a good representation of the heart and a good quality of triangles (here one wants preserve isotropy in R3).
Using interfaces provided by dtk, the pipeline therefore resorts to algorithms from the ITK library (https://itk.org/) for the medical imaging and algorithms from Cgal library (http://www.cgal.org/) for the geometry.
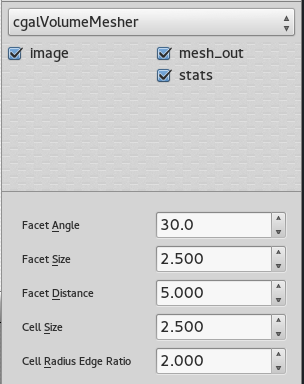
Volume mesher
The mesher used in this case is the Cgal volume mesher, which produces good quality elements. The size of these elements can be easily modified through the interface of dtk-visual-programming, as well as the different properties of the mesh.
 |
 |
|---|
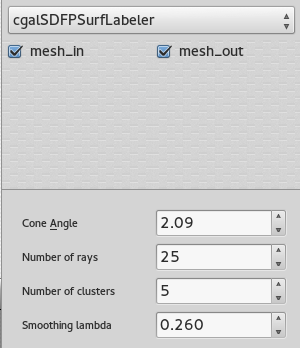
SUrface extraction and labelling
Once the volume is tessellated, the tool Mesh extract surfaces is invoked to recover only the external surface (possibly external surfaces). This surface goes through the Polyhedral labeler, in order to be segmented into an appropriate number of connex components, based on its volume properties. For more information on the different parameters available, consider reading : Consistent mesh skeletonisation.
The number of clusters fixed in the GUI is an approximation of the final number of parts.
Since the CGAL mesher works as a probe slides_L3 (which intersects the appropriate level set, f(x,y,z)=0 for a binary image), it is appropriate to produce multi resolution surfaces of the same heart. It will be possible using the CGAL library, to keep track of the distance between the high resolution model and the lower resolution model.
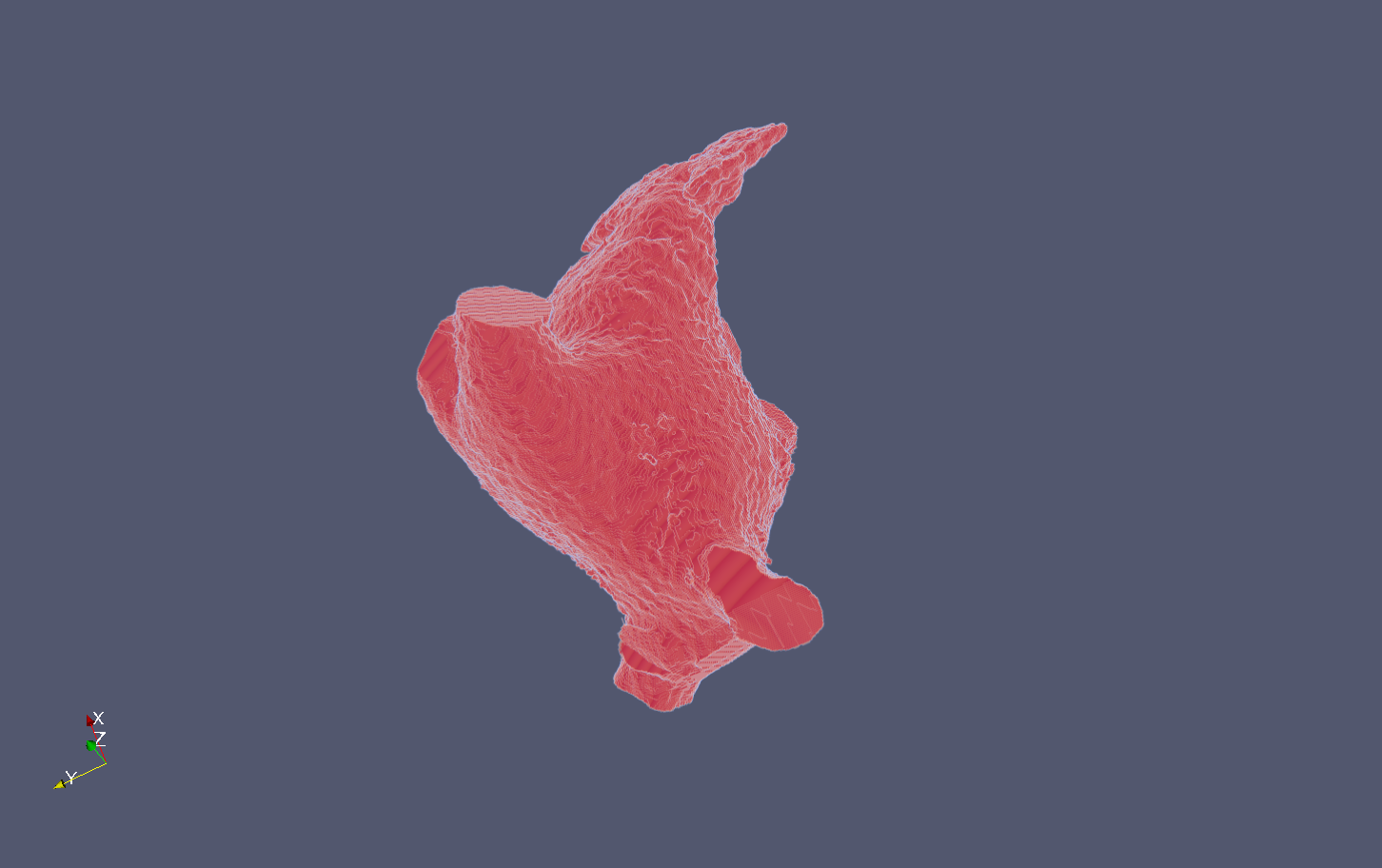
Results
The following snapshots display the initial image (.mha) and the result of the process (.vtu) :