What is dtk?
When we carry out software development in Inria research teams, recurring questions arise:
- “We have lots of software librairies and applications, that are all equally important but we cannot maintain them. Besides, we do not know where to add new features.”
- “We need a modular architecture, but how to do it?”
- “We want to work with another Inria projet team (transversal development) but we do not share standards.”
- “We want to use our historical C/C++ libraries with python/C#/java/matlab/…”
- “We need a crossplatform build system”
- “We need a GUI”
- “We need guidelines”
Such questions point out some deficiencies in research codes:
- lack of architecture design. In practice, a research code is the sum of very heterogeneous contributions coming from PhD students, postdoctoral fellows, and third-party libraries. The end result is a typical spaghetti code.
- lack of infrastructure for the development cycle of software: no version control system, handmade build system, no tool for packaging, publishing or collaborating. A direct consequence is a misallocation of development time: 80% of the effort are placed in areas that do not relate to business code.
- lack of use of standards (design patterns, object oriented concepts). It follows that it is difficult to integrate newcomers and build software community. It is also very tricky to insert new functionalities without breaking everything (which explain the big numbers of forks).
dtk is an attempt to address these issues. It has been devised as a meta-platform providing key ingredients to design scientific modular platforms for Inria research teams. In this context, the framework must enable to:
- encapsulate C/C++ and Fortran codes
- ensure code efficiency (compilation, multi-threading, distributed calculations)
- ensure consistency, evolution and publication of scientific standards for the research teams
These requirements explains why we did not resort to standard Java tools such as Eclipse.
A dtk-based platform features:
- modular architecture
- modular use
- cross-OS build system
- consistent programming foundations
- consistent project management dynamic
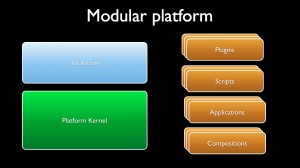
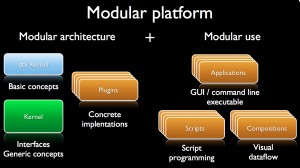
Modular platform
A modular platform first relies on a modular architecture providing several software bricks but also on the possibility to use and interchange these bricks in a very flexible way so as to design efficient workflows for the given scientific fields.
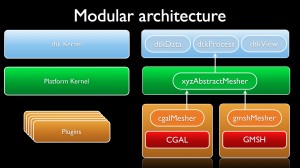
Modular architecture
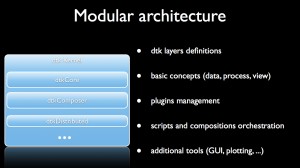
Every dtk-based platform features a generic kernel, an applicative kernel dedicated to the scientific field and a set of atomic software components or plugins.
The generic kernel features several software layers where the basic concepts are defined (typically data, process and view) and where additional tools are provided such as:
- the plugin management system and the factories
- editors to write scripts in different languages (python, tcl , C#, scilab)
- composition workspace to design visual programs that assemble plugins
- a controller/server/slave architecture to submit computation on Inria clusters
- tools for GUI and plotting
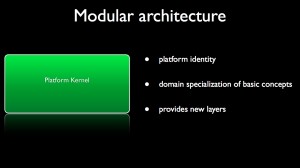
The applicative kernel gives an identity to the platform in relation with the addressed scientific field. From the three basic concepts of the generic kernel, one can devise interfaces defining fundamental concepts of the scientific domain. For instance, in geometry, data would be specialized into spline or mesh; in medical imaging, data would be specialized into image while in numerical simulation, process would be specialized as numerical schemes. Moreover, the applicative kernel can provide additional layers for dealing with data base for instance.
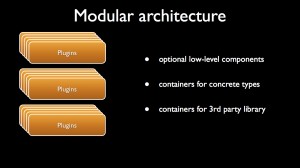
When the interfaces of basic scientific concepts are defined, one can implement them through plugins that are containers of concrete types. These plugins can embed third-party library if needed.
Finally, the principles of a modular architecture can be summarized as follows:
- From one of the three basic dtk concepts (data, process, view), one normalizes an interface of a scientific concept into the kernel of the platform. For instance, a mesher can be defined, especially its inputs and its outputs;
- Then, one implements several meshers into different plugins using if needed third party libraries. In the present case, one implementation makes the use of CGAL when the other wraps GMSH. These two plugins are interchangeable inside the client code thanks to the common interface of the mesher defined in the platform.
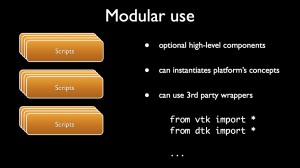
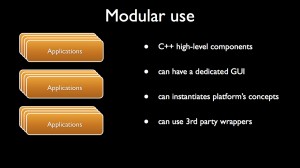
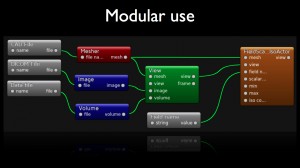
Modular use
A dtk based platform provides several ways to define chains of treatment (aka workflows) that combine plugins:
- via scripts;
- via applications (command line or GUI);
- via visual programming framework.
A classical way to handle the concepts provided by the platform is to use scripts that can be written in python, C# or Tcl for instance. One can make high-level software components that can embed third-party wrappers.
Another mean to use the platform is to resort to plain C++ code to create executables that will be handled via command line or ad hoc GUI.
Eventually, dtk provides a visual programming framework that enables users to connect according to their inputs/outputs, the scientific concepts embedded into visual nodes in order to draw workflows. The resulting compositions can be easily given to co-workers. One can also make sub-compositions that can be inserted into larger ones. The main advantage of this tool is that it does not required the kwowledge of an additional programming language.
Findings
A platform is said modular when it both provides a modular architecture and a modular use.
dtk-based platforms
Today, several software engineering projects are being developed using dtk architecture leading to specific platforms dedicated to very different scientific fields:
- num3sis
- axel
- medinria
- carbonquant
- enas
- in@lgae
- pib
- winpos
- sup
- fsd3d++
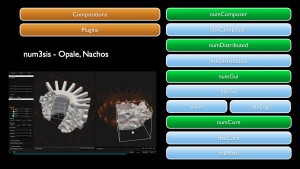
num3sis
The platform num3sis developed by EPI ACUMES (former OPALE) and NACHOS is devoted to scientific computing and numerical simulation. It is not restricted to a particular application field, but is designed to host complex multidisciplinary simulations. Main application fields are currently Computational Fluid Dynamics (CFD), Computational Structural Mechanics (CSM), Computational Electro-Magnetics (CEM). Nevertheless, num3sis can be a framework for the development of other numerical simulation tools for new application areas, such as pedestrian traffic modeling for instance.
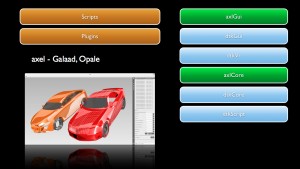
Axel
Axel is an algebraic geometric modeler developed by EPI GALAAD that aims at providing “algebraic modeling” tools for the manipulation and computation with curves, surfaces or volumes described by semi-algebraic representations. These include parametric and implicit representations of geometric objects. Axel also provides algorithms to compute intersection points or curves, singularities of algebraic curves or surfaces, certified topology of curves and surfaces, etc. A plugin mechanism allows to extend easily the data types and functions available in the plateform.
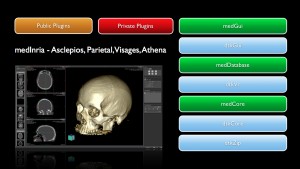
medInria
medInria is a multi-platform medical image processing and visualization software developed by EPI Asclepios, Athena and Visage. It is free and open-source. Through an intuitive user interface, medInria offers from standard to cutting-edge processing functionalities for your medical images such as 2D/3D/4D image visualization, image registration, diffusion MR processing and tractography.
CarbonQuant
CarbonQuant software developed by EPI TOSCA aims at pricing carbon dioxyd gas discharge for the european market. The carbon market was launched in the European Union in 2005 as part of the EU’s initiative to reduce its greenhouse gas (GHG) emissions. For industrial actors, this means that they now have to include their GHG emissions in their production costs. CarbonQuant aims at modeling the behavior of an industrial agent who faces uncertainties, and it compares the expected value of optimal production strategy when buying (or not buying) supplementary emission allowances. This allows us to compute the agent’s carbon indifference price. The resulting indifference price is used to study carbon market sensitivity with respect to the shape of the penalty and the emission allowances allocation.
EnaS
EnaS developed by EPI NeuroMathComp enables analysis of neural population in large scale spiking networks. With the advent of new Multi-Electrode Arrays techniques (MEA), the simultaneous recording of the activity up to hundreds of neurons over a dense configuration supplies today a critical database to unravel the role of specific neural assemblies. Thus, the analysis of spike trains obtained from in vivo or in vitro experimental data requires suitable statistical models and computational tools. The EnaS software offers new computational methods of spike train statistics. It also features several statistical model choices and allows a quantitative comparison between them. Eventually, it provides a control of finite-size sampling effects inherent to empirical statistics.
In@lgae
In@lgae is jointly developed by EPI Biocore and Ange. Its objective is to simulate the productivity of a microalgae production system, taking into account both the process type and its location and time of the year. A first module (Freshkiss) developed by Ange computes the hydrodynamics, and reconstructs the Lagrangian trajectories perceived by the cells. Coupled with the Han model, it results in the computation of an overall photosynthesis yield. A second module is coupled with a GIS (geographic information system) to take into account the meteorology of the considered area (any location on earth). The evolution of the temperature in the culture medium together with the solar flux is then computed. Finally, the productivity in terms of biomass, lipids, pigments together with CO2, nutrients, water consumption, … are assessed. The productivity map which is produced can then be coupled with a resource map describing the availability in CO2 nutrients and land.
Biological Image Platform (PIB)
The PIB platform developed by EPI Morphem provides visual programming framework for the definition of workflows that enable to characterize and model the development and the morphological properties of biological structures from the cell to the supra-cellular scale.
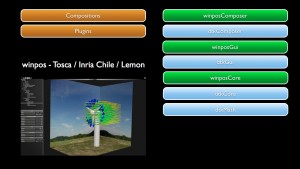
Winpos
Winpos software is being developed in the context of the French-Chilean Associated Team ANESTOC-TOSCA involving the CIRIC at Inria Chile. This project aims at transfering and valuing to Chilean companies the results of researches on renewable energies, mainly wind prediction at the windfarm’s scale by developing and improving the Winpos software based on the downscaling methods, and waves energy potential of a site using video and developping stochastic models for the Wave Energy Converter called Oscillating Water Column.
Scene Understanding Platform (SUP)
SUP is developed by the EPI STARS for perceiving, analysing and interpreting a 3D dynamic scene observed through a network of sensors. SUP´s primary goal is the dissemination of components containing algorithms developed by members of Stars. The dissemination is targeted for use in real-world applications requiring high-throughput.
FS3D++
FS3D++ is a platform developed by EPI APICS, Athena and by the CMA of Mines-Paritech. It is dedicated to the resolution of inverse source problems in electroencephalogrphy (EEG). From pointwise measurements of the electric potential, numerically obtained or taken by electrodes on the scalp, FS3D++ estimates pointwise dipolar current sources within the brain.